With the advent of new collaborative multi-centre research projects, the necessity of having native data format that is flexible, robust and cross-platform compatible became a priority. With this in mind a new file format for ICM+ capable of answering all these needs was created. It is based on the HDF5 format and is supported by most of the data analysis tools available to science.
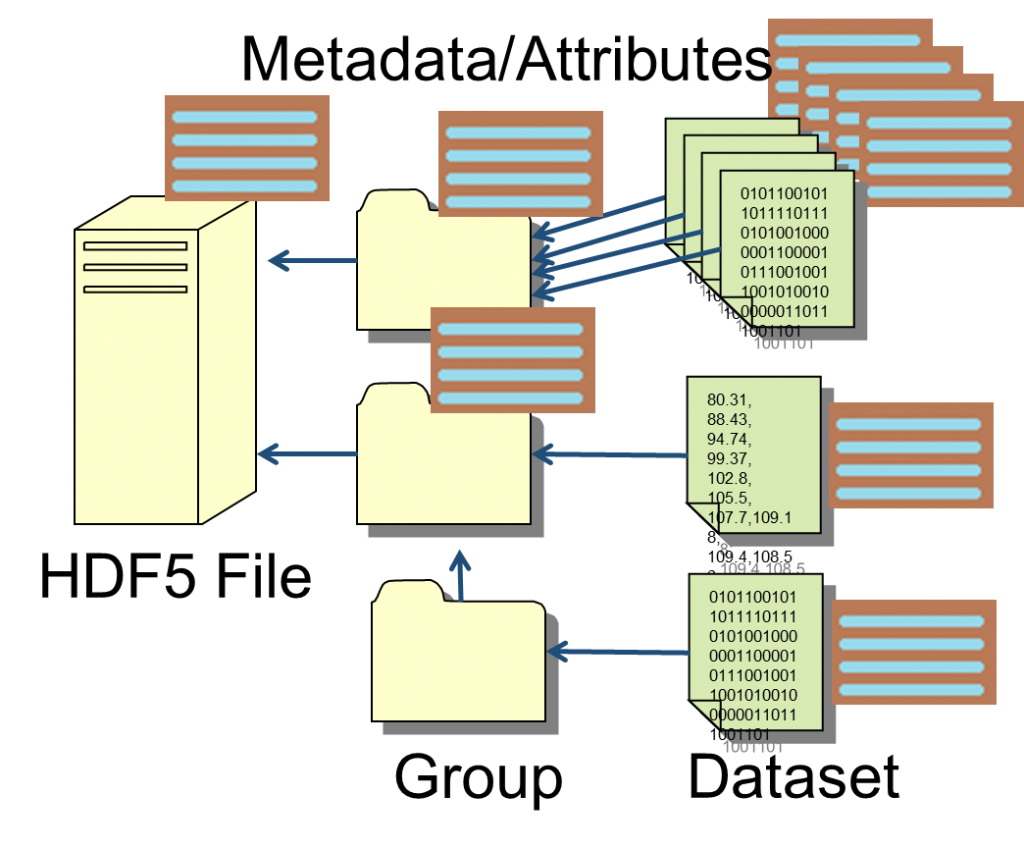
An HDF5 file is composed of 2 main building blocks: Groups and Datasets. The groups form the tree’s stem and branches and are therefore responsible for the hierarchical organisation of the data. Every dataset is a uniform multidimensional array of data objects or elements, represented by one of the predefined simple data-types or compound elements composed of a mixture of data-types. Groups and datasets can be further annotated with metadata contained in associated ‘Attributes’. View image
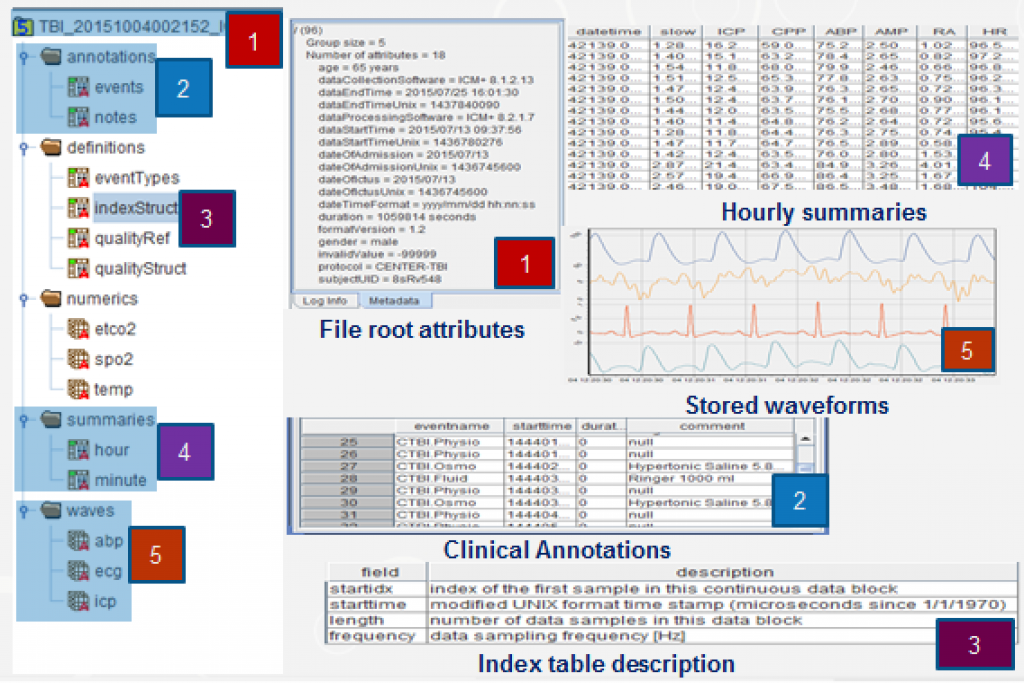
The data is divided in different groups (categories), that are represented as folders in the picture. Each group contains a specific data type associated with it and a set of attributes.
Waveform (5) and numeric data sets include one data set per modality or, in case of composite recordings such as EEG, one group of datasets (individual channels) per modality.
Each data set in turns includes a series of uninterrupted (continuous) data streams characterised by its position in the data set, its actual start time, and its sampling frequency detailed in the dataset ‘Index Table’ attributes (3). In addition each of those datasets also has an attribute for data quality tracking. The annotation group (2) will contain data related to interventions on the patients, where each entry will have its own time stamp, and a quality flag where relevant.
In addition, the file structure also contains: Summaries (4) for calculated data at low sampling frequencies.
Finally, a set of attributes was chosen for the root group containing the most important metadata describing the datasets (1). View image

